Base EditingUnderstanding the human genome
The human genome has four types of bases, which are found in DNA: adenine (A), cytosine (C), guanine (G), and thymine (T). These bases are the “letters” that spell the genetic code that holds the instructions of life.
The genome comprises more than 3 billion of these base pairs in 2 intertwining double strands of DNA; the sequence of these bases encodes genes. Misspellings of even a single letter of a gene, known as a point mutation, can result in disease.
Many human genetic diseases are due to point mutations. In fact, amongst the over 50,000 human disease-causing variants described in a mutation database, about 30,000 are point mutations.
Understanding base editing
Beam has a deep portfolio of proprietary gene editing tools in active use by us and our partners, including base editing, prime editing and nuclease editing. Our current base editor technology includes a wide range of cytosine and adenine base editors, reflecting years of investment and learning in the optimal design and application of these powerful next-generation editing tools.

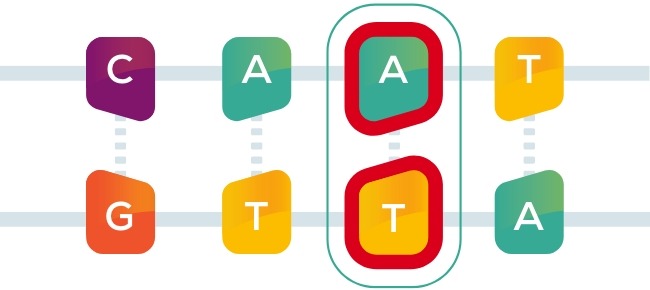
C-to-T base editor chemically modifies cytosine
(C) to thymine (T)

A-to-G base editor chemically modifies
adenine (A) to guanine (G)
Base editors have two principal components:
A CRISPR protein, bound to a guide RNA, that leverages the established DNA-targeting ability of CRISPR, but modified to not cause a double-stranded break.
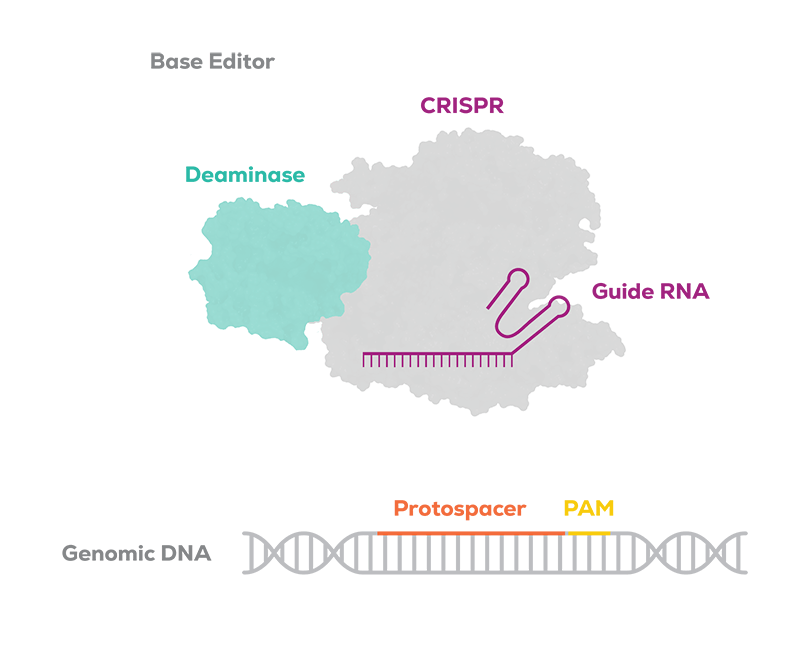
A base editing enzyme, such as a deaminase, which carries out the desired chemical modification of the target DNA base.
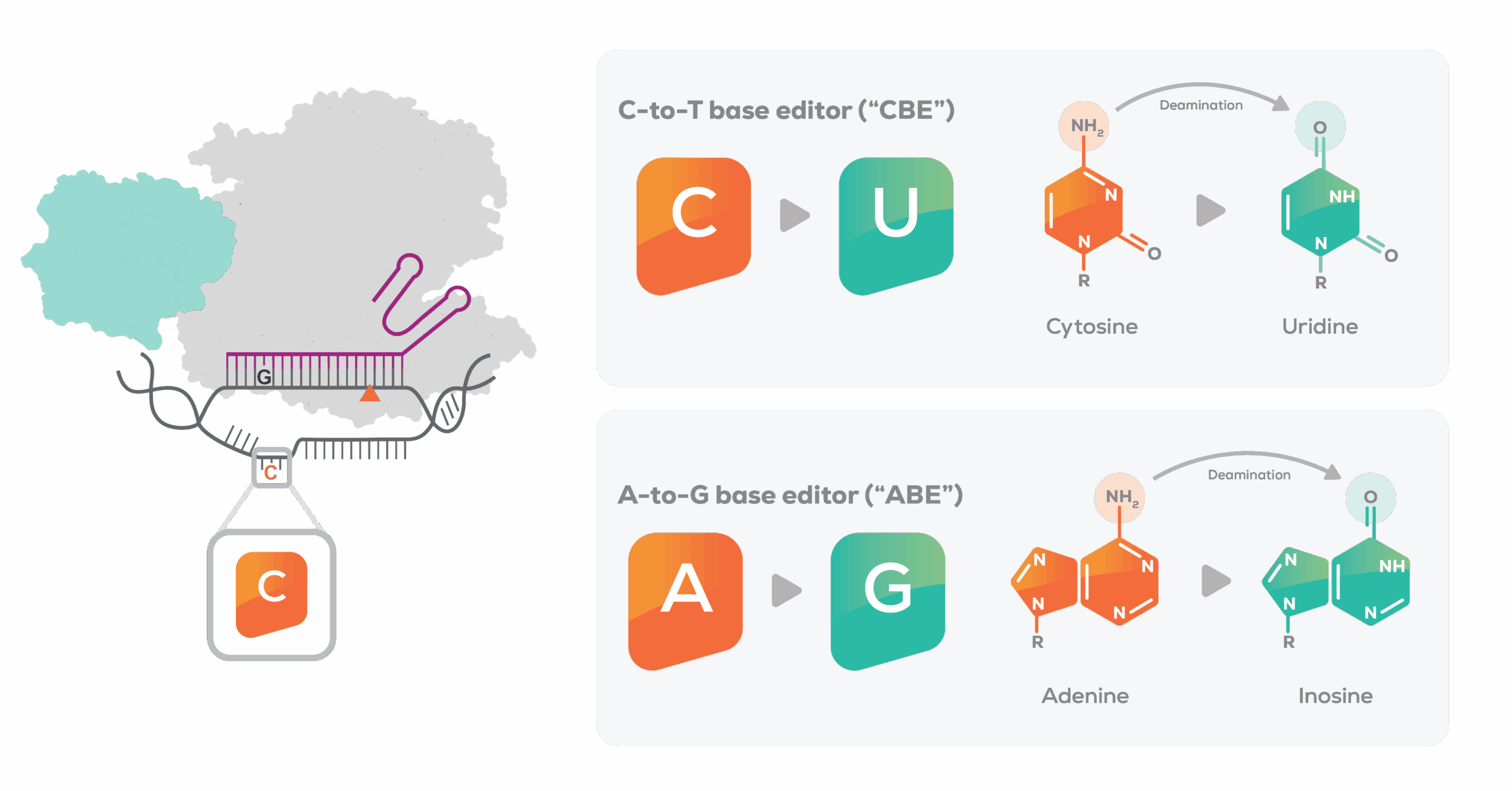
This proprietary combination is designed to enable the precise targeting and editing of a single base pair of DNA or multiple bases at the same time. When introduced into a cell, the CRISPR protein targets the desired genomic location by recognizing a complementary section on the DNA to the section encoded in the guide RNA. The base editor binds the target DNA and exposes a narrow editing window. The deaminase then makes the desired edit to a target base.
We believe that the modular and individual components of our base editors can be customized for specific diseases and create new therapeutic programs. By changing the guide RNA and/or engineering the CRISPR protein, we can efficiently retarget base editors to different genomic locations based on their gene sequences. By changing the deaminase, we can control which base is edited (eg, C or A).
Potential advantages of base editing
Base editing is an emerging class of investigational precision genetic medicines designed to overcome the limitations of existing approaches and expand the potential of genetic medicine.
We believe base editors have several potential advantages over existing gene editing approaches:
1
The creation of precise, predictable, and efficient genetic outcomes at a targeted sequence
2
High-efficiency editing in both dividing and nondividing cells without the need for template-based, homology-directed repair
3
Avoidance of the unwanted consequences associated with double-stranded DNA breaks, such as unpredictable insertions and deletions at the break site and larger-scale genomic rearrangements
Base Editing Applications
Our delivery strategy for base editing includes a suite of clinically validated technologies, including electroporation and nonviral delivery modalities.

